10X Genomics Chromium Single-Cell System
The 10X Genomics Chromium Single-Cell System is used to provide a single-cell expression profiling technology that allows for high-throughput single cell transcriptomics of many different cell types as well as single-nuclei expression profiling. The flexible workflow encapsulates 100 to 10,000 cells or nuclei per library together with micro-beads into nano-droplets. Each bead is loaded with adapters containing one of 750,000 different barcodes for the single cell RNA-seq library preps. Up to eight samples can be processed per batch. The resulting data can be analyzed with the free Cell Ranger and Loupe Cell Browser software. In addition the IIHG Bioinformatics Division has developed a custom single-cell data analysis pipeline for 10X data.
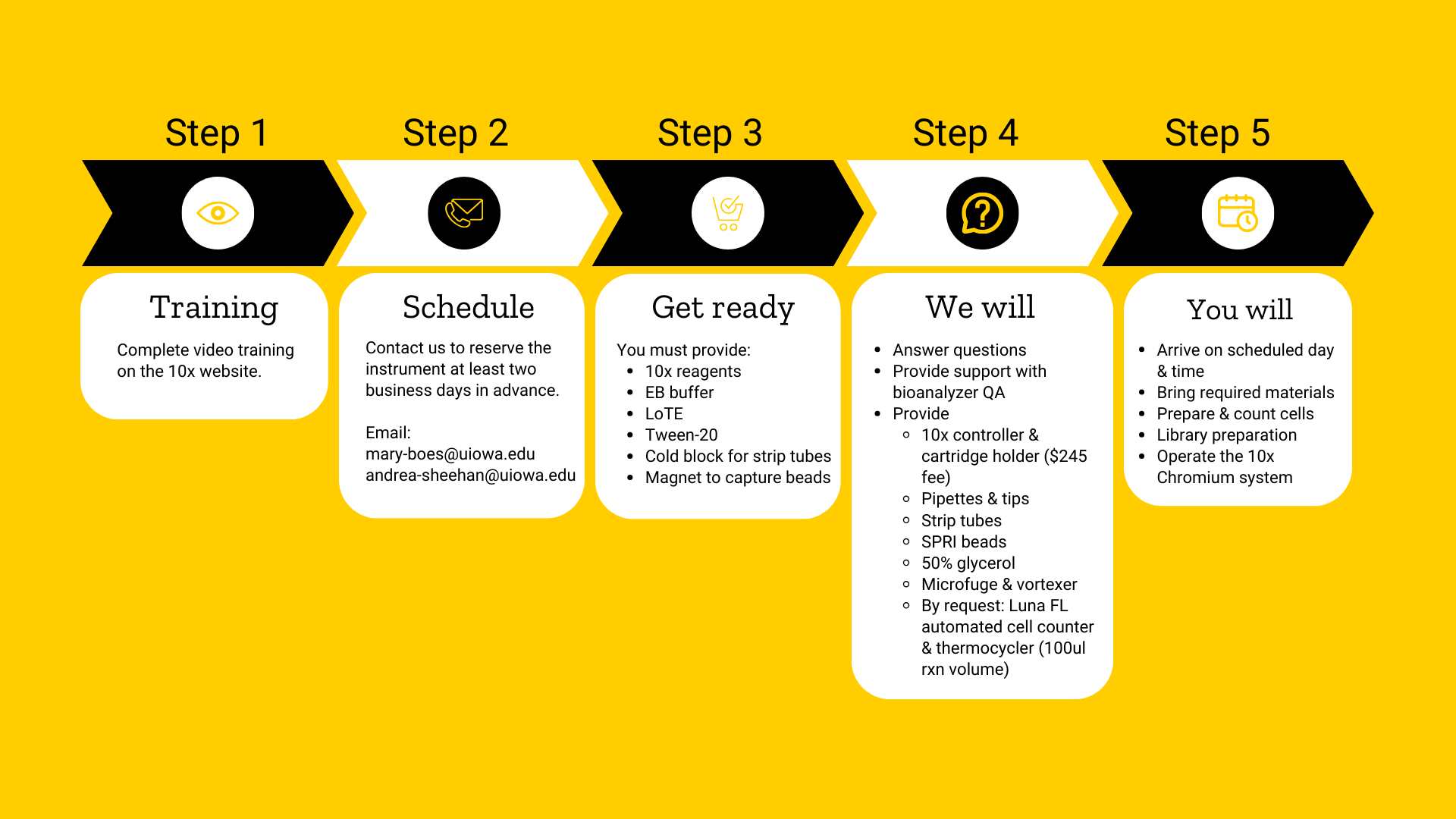
The service operates the 10X Genomics Chromium X and iX Controllers. These controllers support Single-Cell Gene Expression, Immune Profiling, ATAC, and other assays that are supported by the Chromium X controllers. These assays include the measurement of gene expression, cell surface proteins, immune clonotype, antigen specificity, and/or chromatin accessibility.
Fee Schedule
10X Chromium Single Cell Features:
- Fast workflow from cell suspension to 3′ or 5’-cDNA libraries, and T or B cell V(D)J analyses.
- Captures 100-80,000+ cells (from up to 8 samples) in < 7 minutes.
- Recovers up to ~65% of cells (typically 50%).
- Low doublet rate (~0.9% per 1,000 cells).
- Compatible with Element AVITI and Illumina NovaSeq sequencing platforms.
- At least 20,000 reads per cell (or nuclei) should be targeted for sequencing.
- It is possible to work with cryo-preserved cells, enabling safe sample shipping and batching.
- The 10X Genomics-supported cell size limit is 30µm.
- In addition to cell suspension samples, nuclei suspensions can also be used. For information regarding sample preparation, please refer to the 10X cell preparation guide.